Symmetrically dividing bacteria need to place the division machinery in the middle in order to give rise, after cytokinesis, to two equally sized daughter cells. This is achieved by localizing at mid-cell the protein FtsZ, the bacterial tubulin homologue that is needed to assemble a functional septum. A strategy to restrict formation of the FtsZ ring at mid-cell is to have an inhibitor of FtsZ everywhere else but mid-cell. In E. coli this is achieved dynamically with the pole-to-pole oscillations of the Min system.
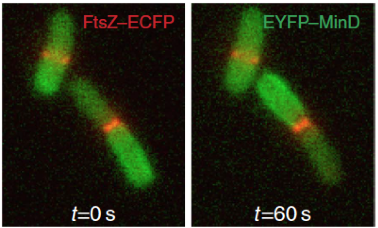
The E. coli Min system comprises only three proteins: MinC, which is the direct inhibitor of FtsZ; MinD, an ATPase that binds to the cytoplasmic membrane; and MinE, that stimulates MinD ATPase activity and triggers the release of MinD from the membrane. Min oscillations emerge from the interplay between the ATP-dependent membrane association and subsequent oligomerization of MinD, and MinE-stimulated local release of MinD from the membrane upon ATP hydrolysis. MinC is a “passive rider” of the oscillations that are exclusively generated by MinD, MinE and the cytoplasmic membrane.
Oscillations have typically a period of 30-60 seconds, therefore each pole is devoid of MinD – and, consequently, MinC – only for a short period of time. Averaging over time, the cell senses an intracellular gradient of MinC with maxima at the poles and minimum at mid-cell, where the FtsZ ring assembles. Cells deprived of the Min system are viable, but assemble septa also at the poles, giving rise to anucleate mini-cells (from which the terms “mini-cell phenotype” and “Min System” originated).
Role of the Min system in chromosome segregation
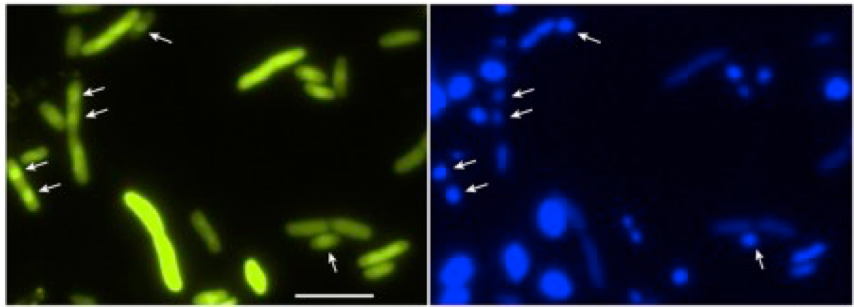
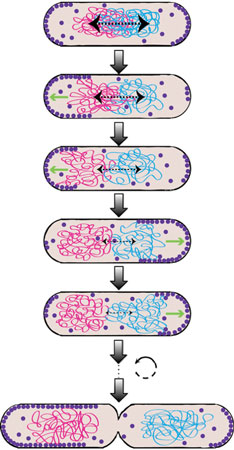
Chromosome segregation is a fundamental process cells have to accomplish, as the genomic DNA encodes for all the protein machineries needed for sustained growth and division. While it is well accepted that eukaryotic cells use an active mechanism for chromosome segregation based on the spindle apparatus, a long-standing debate exists on whether bacterial chromosome segregation is passive or active. We propose that the Min system in E. coli participates in chromosome segregation, thus pushing forward the idea that even small bacterial cells consume energy to segregate their genetic content. We argue that the dynamic gradient of MinD on the membrane ensures at the same time symmetric cell division and proper segregation of the daughter chromosomes. By performing several in vitro and in vivo assays we found that MinD binds to the DNA in a non-sequence specific manner and tethers it transiently to the membrane. Using a mutant MinD (lacking the C-terminal amphipathic helix and having two ariginines mutated to glutamic acid), it is possible to observe localization to the chromosome in E. coli cells.
We think that, by repeated binding and unbinding to MinD, the daughter chromosomes are prevented from moving backwards towards mid-cell and are positioned with high precision and stably away from mid-cell, where cytokinesis can occur. Our model considers that, due to the oscillatory nature of the gradient, each chromosome is biased in its movement toward the pole only during one half of the oscillation cycle, when MinD is mostly at the corresponding pole. During another half of the Min cycle, this chromosome is in principle freely diffusing, but because of the large size of the chromosome its diffusion is much slower than the cycle of Min oscillation and thus it cannot randomize its position on this time scale. While we have already accumulated enough evidence to put this Brownian ratchet-type segregation mechanism forward, much is left to be discovered on the molecular level!
Publications
C-terminal eYFP fusion impairs Escherichia coli MinE function
N. Palanisamy, M. A. Öztürk, E. B. Akmeric and B. Di Ventura
Open Biology, 2020
doi:10.1098/rsob.200010
Chromosome segregation by the Escherichia coli Min system
B. Di Ventura, B. Knecht, H. Andreas, W. J. Godinez, M. Fritsche, K. Rohr, W. Nickel, D. W. Heermann, V. Sourjik
Mol Syst Biol, 2013
doi:10.1038/msb.2013.44
Self-organized partitioning of dynamically localized proteins in bacterial cell division
B. Di Ventura, V. Sourjik
Mol Syst Biol, 2011
doi:10.1038/msb.2010.111