Optogenetics is the use of genetically encoded light-inducible switches that are engineered to exert a certain biological function only after absorption of light of a specific wavelength. The greatest advantage of using light as input to control biological processes is the possibility of local activation with down to subcellular resolution. A very promising natural photoreceptor that is being often used in optogenetic tools is the second Light Oxygen Voltage domain of Avena sativa phototropin 1 (AsLOV2). This is a small (12 kDa) soluble domain that undergoes a conformational change when absorbing blue light (450-480 nm), so that its N- and C-terminal helices unfold and undock from the core LOV domain. The AsLOV2 domain – as well as other LOV domains from different organisms – has been successfully fused to other proteins to control their activity via light. Another approach consists in exploiting the light-induced unfolding and undocking of the C-terminal Jα helix of the AsLOV2 domain to control via light the exposure of small peptides that are either appended to or embedded into the Jα helix itself.
Controlling nuclear protein import with LINuS
Cell biology is pervaded with examples of processes that are controlled by the regulated import of proteins into the nucleus. Transcription factors are clear targets of such regulation since, when retained in the cytoplasm, they have no access to the promoters they bind to and are, therefore, kept in a latent form. Other classes of proteins, for instance kinases, whose function is exerted in the nucleus, are also sequestered in the cytoplasm until the appropriate signaling cascade is activated that triggers their relocation to the nuclear compartment. Since it has become clear that proteins can have distinct or even opposing functions in different compartments, the ability to manipulate protein localization in a precise temporal fashion is of paramount importance for dissecting cellular pathways. We have recently developed an optogenetic tool, called LINuS (Light-Inducible Nuclear localization Signal), which allows importing proteins of interested into the nucleus by shining blue light onto the cells.
We have proven that LINuS is functional in yeast, in several mammalian cell lines (HEK 293T, HeLa, HepG2, C2C12, HCT116, H1299, MCF7) and in zebrafish.
 | 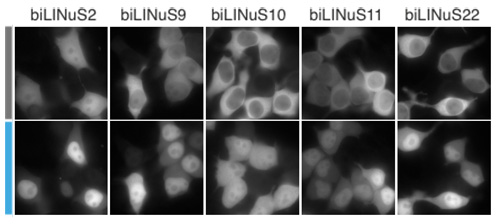 |
Protein nuclear import can be controlled by blue light using LINuS. (left) Schematic of the rationale behind LINuS. In the dark state, the hybrid Jα helix is folded and interacts with the AsLOV2 core domain. After blue light exposure, it is in an unfolded and undocked state, thus the NLS is accessible to endogenous importins. (right) Representative fluorescence microscopy images of HEK 293T cells transiently transfected with the indicated mCherry-biLINuS variants before (grey bar) and after (cyan bar) 15 min of illumination. All biLINuS variants carry a constitutive PKIt NES.
Controlling nuclear protein export with LEXY
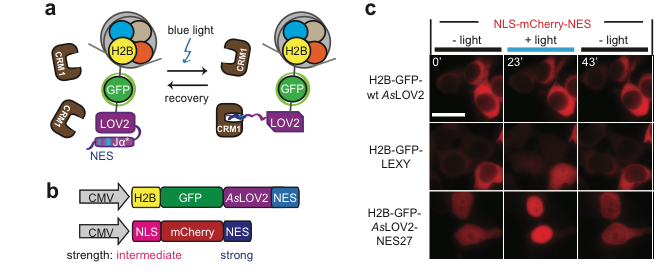
Following a similar principle, we have engineered LEXY (Light-inducible EXport sYstem) to control with blue light nuclear protein export. In this case, we have introduced mutations into the Jα helix of the AsLOV2 domain to include residues essential for export function. LEXY is as flexible as LINuS, working with different proteins of interest and in different cell types.
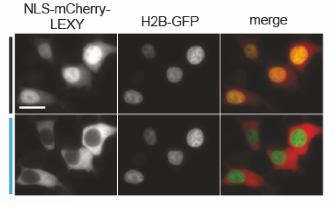
By fusing LEXY to the very abundant histone H2B, we created a light-inducible Leptomycin B treatment to block endogenous protein export.
Publications
Engineering AraC to make it responsive to light instead of arabinose
E. Romano, A. Baumschlager, E. B. Akmeriç, N. Palanisamy, M. Houmani, G. Schmidt, M. A. Öztürk, L. Ernst, M. Khammash and B. Di Ventura Nature Chemical Biology, 2021
doi:10.1101/2020.07.14.202911
Optogenetic Control of Nuclear Protein Import in Living Cells Using Light-Inducible Nuclear Localization Signals (LINuS)
P. Wehler, D. Niopek, R. Eils, B. Di Ventura
Curr Protoc Chem Biol, 2016
doi:10.1002/cpch.4
Optogenetic control of nuclear protein export
D. Niopek, P. Wehler, J. Roensch, R. Eils, B. Di Ventura
Nat Commun, 2016
doi:10.1038/ncomms10624
Go in! Go out! Inducible control of nuclear localization
B. Di Ventura, B. Kuhlman
Curr Opin Chem Biol, 2016
doi:10.1016/j.cbpa.2016.06.009
Engineering light-inducible nuclear localization signals for precise spatiotemporal control of protein dynamics in living cells
D. Niopek, D. Benzinger, J. Roensch, T. Draebing, P. Wehler, R. Eils, B. Di Ventura
Nat Commun, 2014
doi:10.1038/ncomms5404